Project 2789: A. Bignon, B. G. Waisfeld, N. E. Vaccari, B. D. Chatterton. 2020. Reassessment of the Order Trinucleida (Trilobita). Journal of Systematic Palaeontology. 18 (13):1061-1077.
Abstract
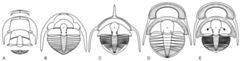
Trinucleoids have been treated as a superfamily of the Order Asaphida, based on: a pre-occipital glabellar tubercle, a cephalic median suture in most ancestral forms and the presence of broadly similar globular protaspid larvae in the life cycle. Recent discoveries cast doubts on the monophyly of the two last characters. This study provides a phylogenetic analysis whose main purpose is to test the affiliation of trinucleoids. The topology of the strict consensus tree obtained indicates that trinucleoids share a more recent common ancestor with members of the Order Ptychopariida than they do with any asaphid. The stratigraphically higher and most derived trinucleoids are well defined and easy to recognize. Their basal representatives can also be identified with confidence, in spite of substantial variation in the diagnostic characters of the group. These early trinucleoids are similar enough to the typical basal “ptychopariid” bauplan to suggest ancestry within that diffuse and poorly defined trilobite order. Indeed, Ptychopariida is presently regarded as being paraphyletic. We consider that raising trinucleids to ordinal status, by proposing the Order Trinucleida, will make it easier to decipher higher level relationships among trilobites. This action, sustained by improved morphologic and ontogenetic information of Cambro-Ordovician trilobites, reveals convergences of the three characters that have been used to support the inclusion of trinucleids within Asaphida. Based on this new information, we propose six distinct morphotypes of commutavi protaspis and, as a probable sensorial organ, that the cephalic axial tubercle migration might be caused by an increase in the height of the glabella and thus may not bear an important systematic signal. Moreover, the tree topology provided herein offers a preliminary overview of evolution within the order. Alsataspididae and Raphiophoridae are paraphyletic, suggesting that more investigation is needed to define them clearly. Dionidae appears to be more closely related to Raphiophoridae than to Trinucleidae, suggesting that the character state of ‘a perforated fringe’ evolved more than once.Read the article »
Article DOI: 10.1080/14772019.2020.1720324
Project DOI: 10.7934/P2789, http://dx.doi.org/10.7934/P2789
| This project contains | Matrices |
|---|---|
Download Project SDD File | Total scored cells: 4661 Total media associated with cells: 0 Total labels associated with cell media: 0 |
| Characters | |
| Total characters: 75 Total characters with associated media: 14 Total characters with media with labels: 0 Total character states: 185 Total character states with associated media: 130 Total character states with media with labels:0 Total unordered/ordered characters:75/0 |
Currently Viewing:
MorphoBank Project 2789
MorphoBank Project 2789
- Creation Date:
22 November 2017 - Publication Date:
31 December 2019 - Media downloads: 7

- Matrix downloads: 16

Authors' Institutions ![]()
- Universidad Nacional de Cordoba (UNC)
- University of Alberta
- Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)
- Universidad Nacional de la Rioja
Members
| member name | taxa |
specimens |
media | chars | character
| cell scorings (scored, NPA, "-") | cell
| rules | ||||||
| Arnaud Bignon Project Administrator | 91 | 3 | 151 | 75 | 144 | 0 | 4661 (4276, 0, 385) | 0 | 0 | 0 | ||||
| Brian Chatterton Full membership | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0, 0, 0) | 0 | 0 | 0 | ||||
| Emilio Vaccari Full membership | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0, 0, 0) | 0 | 0 | 0 | ||||
| Beatriz Waisfeld Full membership | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0, 0, 0) | 0 | 0 | 0 | ||||
Taxonomic Overview for Matrix 'M24906' (72 Taxa)
| taxon | unscored cells |
scored cells |
no cell support |
NPA cells |
"-" cells | cell images | labels on cell images |
member access |
| [1] † Myinda uriconii Taxon name last Modified on 08/28/19 | 37 | 36 | 36 | 0 | 2 | 0 | 0 | 4 |
| [2] † Myindella crux Taxon name last Modified on 08/28/19 | 6 | 67 | 67 | 0 | 2 | 0 | 0 | 4 |
| [3] † Hanchungolithus richthofeni Taxon name last Modified on 11/29/18 | 5 | 68 | 68 | 0 | 2 | 0 | 0 | 4 |
| [4] † Ningkianolithus welleri Taxon name last Modified on 08/28/19 | 32 | 41 | 41 | 0 | 2 | 0 | 0 | 4 |
| [5] † Reedolithus quebecensis Taxon name last Modified on 12/04/18 | 3 | 70 | 70 | 0 | 2 | 0 | 0 | 4 |
| [6] † Protoincaia ancestor Taxon name last Modified on 12/04/18 | 10 | 56 | 56 | 0 | 9 | 0 | 0 | 4 |
| [7] † Tretaspis ceryx Taxon name last Modified on 12/04/18 | 6 | 60 | 60 | 0 | 9 | 0 | 0 | 4 |
| [8] † Stapeleyella inconstans Taxon name last Modified on 12/04/18 | 11 | 55 | 55 | 0 | 9 | 0 | 0 | 4 |
| [9] † Bancroftolithus hughesi Taxon name last Modified on 12/04/18 | 0 | 73 | 73 | 0 | 2 | 0 | 0 | 4 |
| [10] † Pragolithus praecedens Taxon name last Modified on 11/29/18 | 15 | 51 | 51 | 0 | 9 | 0 | 0 | 4 |
| [11] † Trinucleus fimbriatus Taxon name last Modified on 12/04/18 | 6 | 60 | 60 | 0 | 9 | 0 | 0 | 4 |
| [12] † Cryptolithus tesselatus Taxon name last Modified on 11/29/18 | 0 | 63 | 63 | 0 | 12 | 0 | 0 | 4 |
| [13] † Deanaspis goldfusii Taxon name last Modified on 11/29/18 | 5 | 58 | 58 | 0 | 12 | 0 | 0 | 4 |
| [14] † Onnia s. superba Taxon name last Modified on 11/29/18 | 2 | 61 | 61 | 0 | 12 | 0 | 0 | 4 |
| [15] † Marrolithus bureaui Taxon name last Modified on 11/29/18 | 2 | 61 | 61 | 0 | 12 | 0 | 0 | 4 |
| [16] † Dionide mareki Taxon name last Modified on 11/29/18 | 1 | 62 | 62 | 0 | 12 | 0 | 0 | 4 |
| [17] † Trinucleoides reussi Taxon name last Modified on 08/28/19 | 5 | 58 | 58 | 0 | 12 | 0 | 0 | 4 |
| [18] † Ampyxella edgelli Taxon name last Modified on 11/29/18 | 9 | 55 | 55 | 0 | 11 | 0 | 0 | 4 |
| [19] † Globampyx trinucleoides Taxon name last Modified on 08/28/19 | 3 | 61 | 61 | 0 | 11 | 0 | 0 | 4 |
| [20] † Damghanampyx ginteri Taxon name last Modified on 11/29/18 | 6 | 59 | 59 | 0 | 10 | 0 | 0 | 4 |
| [21] † Raphioampyx sinankylosus Taxon name last Modified on 12/04/18 | 19 | 45 | 45 | 0 | 11 | 0 | 0 | 4 |
| [22] † Rhombampyx glaber Taxon name last Modified on 12/04/18 | 15 | 49 | 49 | 0 | 11 | 0 | 0 | 4 |
| [23] † Raphiophorus parvulus Taxon name last Modified on 12/04/19 | 2 | 62 | 62 | 0 | 11 | 0 | 0 | 4 |
| [24] † Cnemidopyge costatus Taxon name last Modified on 12/08/18 | 14 | 50 | 50 | 0 | 11 | 0 | 0 | 4 |
| [25] † Lonchodomas tenuis Taxon name last Modified on 11/29/18 | 15 | 49 | 49 | 0 | 11 | 0 | 0 | 4 |
| [26] † Ampyx nasutus Taxon name last Modified on 08/28/19 | 18 | 46 | 46 | 0 | 11 | 0 | 0 | 4 |
| [27] † Mendolaspis sanjuaninus Taxon name last Modified on 11/29/18 | 20 | 44 | 44 | 0 | 11 | 0 | 0 | 4 |
| [28] † Nambeetella fitzroyensis Taxon name last Modified on 11/29/18 | 13 | 51 | 51 | 0 | 11 | 0 | 0 | 4 |
| [29] † Pseudampyxina trisegmentata Taxon name last Modified on 12/04/18 | 18 | 46 | 46 | 0 | 11 | 0 | 0 | 4 |
| [30] † Lehnertia nawisapa Taxon name last Modified on 11/29/18 | 2 | 67 | 67 | 0 | 6 | 0 | 0 | 4 |
| [31] † Pyrimetopus pyrifrons Taxon name last Modified on 08/29/19 | 10 | 63 | 63 | 0 | 2 | 0 | 0 | 4 |
| [32] † Orometopus elatifrons Taxon name last Modified on 11/29/18 | 13 | 60 | 60 | 0 | 2 | 0 | 0 | 4 |
| [33] † Araiopleura reticulata Taxon name last Modified on 11/29/18 | 7 | 66 | 66 | 0 | 2 | 0 | 0 | 4 |
| [34] † Hapalopleura clavata Taxon name last Modified on 11/25/17 | 7 | 66 | 66 | 0 | 2 | 0 | 0 | 4 |
| [35] † Seleneceme acuticaudata Taxon name last Modified on 12/04/18 | 25 | 41 | 41 | 0 | 9 | 0 | 0 | 4 |
| [36] † Skljarella cracens Taxon name last Modified on 12/04/18 | 5 | 68 | 68 | 0 | 2 | 0 | 0 | 4 |
| [37] † Joshuaspis parvus Taxon name last Modified on 11/29/18 | 28 | 45 | 45 | 0 | 2 | 0 | 0 | 4 |
| [38] † Doremataspis ornata Taxon name last Modified on 08/28/19 | 14 | 59 | 59 | 0 | 2 | 0 | 0 | 4 |
| [39] † Gibbura lepida Taxon name last Modified on 08/28/19 | 30 | 43 | 43 | 0 | 2 | 0 | 0 | 4 |
| [40] † Liostracina simesi Taxon name last Modified on 11/29/18 | 10 | 63 | 63 | 0 | 2 | 0 | 0 | 4 |
| [41] † Liostracina volens Taxon name last Modified on 11/29/18 | 27 | 46 | 46 | 0 | 2 | 0 | 0 | 4 |
| [42] † Liostracina tangwangzhaiensis Taxon name last Modified on 11/29/18 | 11 | 62 | 62 | 0 | 2 | 0 | 0 | 4 |
| [43] † Nepea narinosa Taxon name last Modified on 08/28/19 | 17 | 56 | 56 | 0 | 2 | 0 | 0 | 4 |
| [44] † Norwoodia boninoi Taxon name last Modified on 08/28/19 | 6 | 67 | 67 | 0 | 2 | 0 | 0 | 4 |
| [45] † Glaphyraspis parva Taxon name last Modified on 08/28/19 | 14 | 59 | 59 | 0 | 2 | 0 | 0 | 4 |
| [46] † Aphelaspis brachyphasis Taxon name last Modified on 11/29/18 | 10 | 63 | 63 | 0 | 2 | 0 | 0 | 4 |
| [47] † Ptychoparia striata Taxon name last Modified on 12/04/18 | 3 | 70 | 70 | 0 | 2 | 0 | 0 | 4 |
| [48] † Elrathia kingi Taxon name last Modified on 11/29/18 | 5 | 68 | 68 | 0 | 2 | 0 | 0 | 4 |
| [49] † Altikolia kurshabica Taxon name last Modified on 08/28/19 | 12 | 61 | 61 | 0 | 2 | 0 | 0 | 4 |
| [50] † Bolaspidella housensis Taxon name last Modified on 08/28/19 | 0 | 73 | 73 | 0 | 2 | 0 | 0 | 4 |
| [51] † Kochaspis liliana Taxon name last Modified on 08/28/19 | 11 | 62 | 62 | 0 | 2 | 0 | 0 | 4 |
| [52] † Gunnia fava Taxon name last Modified on 08/28/19 | 10 | 63 | 63 | 0 | 2 | 0 | 0 | 4 |
| [53] † Modocia whiteleyi Taxon name last Modified on 08/28/19 | 4 | 69 | 69 | 0 | 2 | 0 | 0 | 4 |
| [54] † Hardyoides minor Taxon name last Modified on 11/29/18 | 7 | 66 | 66 | 0 | 2 | 0 | 0 | 4 |
| [55] † Asaphus (A.) expansus Taxon name last Modified on 11/29/18 | 5 | 66 | 66 | 0 | 4 | 0 | 0 | 4 |
| [56] † Megistaspis (m.) limbata Taxon name last Modified on 11/29/18 | 14 | 58 | 58 | 0 | 3 | 0 | 0 | 4 |
| [57] † Asaphellus homfrayi Taxon name last Modified on 08/28/19 | 7 | 65 | 65 | 0 | 3 | 0 | 0 | 4 |
| [58] † Tsinania canens Taxon name last Modified on 08/28/19 | 10 | 62 | 62 | 0 | 3 | 0 | 0 | 4 |
| [59] † Liomegalaspides hupeiensis Taxon name last Modified on 11/29/18 | 4 | 65 | 65 | 0 | 6 | 0 | 0 | 4 |
| [60] † Taihungshania shui Taxon name last Modified on 12/04/18 | 9 | 60 | 60 | 0 | 6 | 0 | 0 | 4 |
| [61] † Nileus armadillo Taxon name last Modified on 08/28/19 | 2 | 68 | 68 | 0 | 5 | 0 | 0 | 4 |
| [62] † Remopleurides xiazhenensis Taxon name last Modified on 12/04/18 | 12 | 57 | 57 | 0 | 6 | 0 | 0 | 4 |
| [63] † Pterocephalia norfordi Taxon name last Modified on 12/04/18 | 2 | 71 | 71 | 0 | 2 | 0 | 0 | 4 |
| [64] † Maladioides coreanicus Taxon name last Modified on 11/29/18 | 10 | 62 | 62 | 0 | 3 | 0 | 0 | 4 |
| [65] † Ivshinaspis reticulata Taxon name last Modified on 11/29/18 | 19 | 53 | 53 | 0 | 3 | 0 | 0 | 4 |
| [66] † Haniwa quadrata Taxon name last Modified on 11/29/18 | 10 | 62 | 62 | 0 | 3 | 0 | 0 | 4 |
| [67] † Abharella sp. Taxon name last Modified on 11/29/18 | 12 | 60 | 60 | 0 | 3 | 0 | 0 | 4 |
| [68] † Ceratopyge forficuloides Taxon name last Modified on 11/29/18 | 16 | 56 | 56 | 0 | 3 | 0 | 0 | 4 |
| [69] † Proceratopyge cf. p. lata Taxon name last Modified on 11/29/18 | 11 | 61 | 61 | 0 | 3 | 0 | 0 | 4 |
| [70] † Parabolina acanthura Taxon name last Modified on 11/29/18 | 4 | 69 | 69 | 0 | 2 | 0 | 0 | 4 |
| [71] † Olenellus gilberti Taxon name last Modified on 08/28/19 | 2 | 70 | 70 | 0 | 3 | 0 | 0 | 4 |
| [72] † Redlichia takooensis Taxon name last Modified on 12/04/18 | 4 | 68 | 68 | 0 | 3 | 0 | 0 | 4 |
Project downloads 
| type | number of downloads | Individual items downloaded (where applicable) |
| Total downloads from project | 166 | |
| Document downloads | 6 | All Trees (4 downloads); Consensus Strict (2 downloads); |
| Project downloads | 137 | |
| Matrix downloads | 16 | Trinucleida Matrix (16 downloads); |
| Media downloads | 7 | M595224 (3 downloads); M595179 (1 download); M595180 (1 download); M595195 (1 download); M466122 (1 download); |