Project 4205: C. Ramírez-Portilla, I. M. Bieger, R. G. Belleman, T. Wilke, J. Flot, A. H. Baird, S. Harii, F. Sinniger, J. A. Kaandorp. 2022. Quantitative three-dimensional morphological analysis supports species discrimination in complex-shaped and taxonomically challenging corals. Frontiers in Marine Science. 9:null.
Abstract
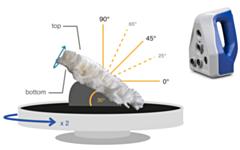
Morphological characters play an important role in species descriptions and are essential for a better understanding of the function, evolution and plasticity of an organism’s shape. However, in complex-shaped organisms lacking characteristic features that can be used as landmarks, quantifying morphological traits, assessing their intra- and interspecific variation, and subsequently delineating phenotypically distinct groups continue to be problematic. For such organisms, three-dimensional morphological analysis might be a promising approach to differentiate morphogroups and potentially aid the delineation of species boundaries, though identifying informative features remains a challenge. Here, we assessed the potential of 3D-based quantitative morphology to delineate a priori and/or to discriminate a posteriori morphogroups of complex-shaped and taxonomically challenging organisms, such as corals from the morphologically diverse genus Acropora. Using three closely related coral taxa previously delimited using other lines of evidence, we extracted a set of variables derived from triangulated polygon meshes and medial axis skeletons of the 3D models. From the resulting data set, univariate and multivariate analyses of 3D-based variables quantifying overall shape including curvature, branching, and complexity were conducted. Finally, informative feature selection was performed to assess the discriminative power of the selected variables. Results revealed significant interspecific differences in the means of a set of 3D-based variables, highlighting potentially informative characters that provide sufficient resolution to discriminate morphogroups congruent with independent species identification based on other lines of evidence. A combination of representative features, remarkably represented by curvature, yielded measures that assisted in differentiating closely related species despite the overall morphospaces overlap. This study shows that a well-justified combination of 3D-based variables can aid species discrimination in complex-shaped organisms such as corals and that feature screening and selection is useful for achieving sufficient resolution to validate species boundaries. Yet, the significant discriminative power displayed by curvature-related variables and their potential link to functional significance need to be explored further. Integrating informative morphological features with other independent lines of evidence appears therefore a promising way to advance not only taxonomy but also our understanding of morphological variation in complex-shaped organisms.Read the article »
Article DOI: 10.3389/fmars.2022.955582
Project DOI: 10.7934/P4205, http://dx.doi.org/10.7934/P4205
| This project contains |
|---|
Download Project SDD File |
Currently Viewing:
MorphoBank Project 4205
MorphoBank Project 4205
- Creation Date:
02 March 2022 - Publication Date:
22 August 2022 - Media downloads: 3

Authors' Institutions ![]()
- Universitaet Giessen (Justus-Liebig-Universität Gießen)
- Université libre de Bruxelles
- Universiteit van Amsterdam (University of Amsterdam)
- James Cook University
- University of the Ryukyus
- Interuniversity Institute of Bioinformatics in Brussels – (IB)2
Members
| member name | taxa |
specimens |
media |
| Catalina Ramirez Portilla Project Administrator | 9 | 74 | 75 |
Project has no matrices defined.
Project downloads 
| type | number of downloads | Individual items downloaded (where applicable) |
| Total downloads from project | 7 | |
| Media downloads | 3 | M840684 (1 download); M840670 (1 download); M840694 (1 download); |
| Project downloads | 4 |